Abstract
Since the 1990s, multiple infectious diseases have “spilled over” from nonhuman animals to infect humans and cause significant global morbidity and mortality. Despite efforts to detect and respond to such threats, surveillance and mitigation efforts have been criticized as ineffective. This article describes what “spillover” and “spillback” events are and canvasses 5 ways in which clinicians can improve emerging microbial pathogen, especially viral, detection and containment responses.
Viral Zoonoses
During the last 25 years, it has been painfully evident that modern medicine, despite all its remarkable advances, is woefully unprepared for epidemic threats, especially viral zoonotic threats.1,2 One need only examine the recent human morbidity from epidemics of West Nile virus (2002-present),3 SARS-CoV-1 (2002-2003),4 Marburg virus (2004-2005),5 H1N1 influenza A pandemic virus (2009-2010),6 MERS-CoV (2012-2019),7 H7N9 avian influenza virus (2017),8 ebolavirus (2013-2016),9 Zika virus (2013-17),10 yellow fever virus (2016-2017),11 Lassa virus (2018),12 and SARS-CoV-2 (2019-present)13 to affirm this position. Part of the difficulty in preventing such zoonotic viral epidemics is a lack of understanding of the nature of these threats. Here, we summarize observations on zoonotic viral spillover and spillback that we believe every clinician should know. Having this knowledge will aid clinicians in sounding a public health alarm should they be among the first to witness a new zoonotic emerging infectious disease problem.
Definitions
The following concepts are key to understanding the nature and scope of zoonotic threats.
- Infection. Porta et al define infection as “the entry and development or multiplication of an infectious agent in an organism, including the body of humans and animals.”14 Infections do not always cause signs and symptoms of disease.
- Zoonosis. Any disease that can be transmitted from nonhuman animals to humans, or from humans to nonhuman animals (reverse zoonosis).15
- Emerging infection disease. An infectious disease that has newly appeared in a population or has existed previously but is rapidly spreading in terms of either the number of people infected or the geographical areas affected.16
- Spillover. Microbe transmission “from one species (usually the reservoir but potentially an amplifying or bridge host) to a novel, susceptible species, establishing infection in this individual new host”17 (see Figure 1).
- Spillback. A specific case of spillover when a microbe spills from a new host back to the original host (see Figure 1).17
- One Health. The Association Veterinary Medical Association defines One Health as “The integrative effort of multiple disciplines working locally, nationally, and globally to attain optimal health for people, animals, and the environment.”18
Figure 1. Virus Spillover and Spillback
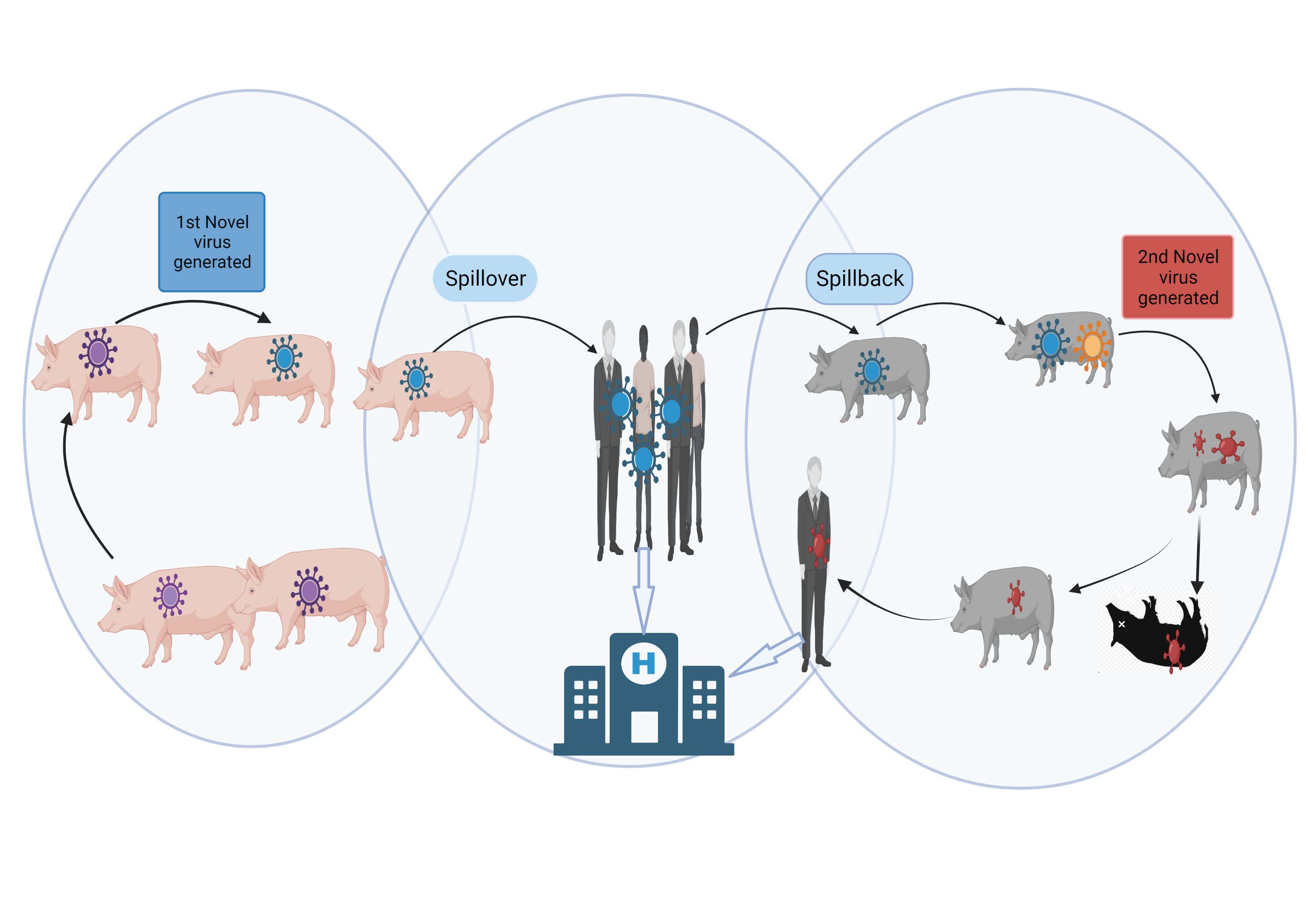
Created with BioRender.com.
Abbreviation: H, human hospital.
A novel virus (blue virus) emerges in pigs and is transmitted to humans in a spillover event. This virus is amplified in humans and spills back to another group of pigs. In this second group of pigs, the virus mixes with other swine viruses and changes yet again (red virus). This new virus has the potential to cause new morbidity in both pigs and humans.
Lesson 1: Viruses Are Everywhere
We are continually challenged by viruses from both outside and within our bodies. Viruses of many kinds surround and threaten us, yet relatively few have the capacity to cause disease (see Figure 2). Exact counts of unique viruses or measurements of total viral mass are not available, but it is agreed that viruses greatly outnumber bacteria and that viruses can infect all forms of life, including bacteria and single cell organisms. It has been estimated that there are more than 10 nonillion (1030) individual virus particles on earth,19,20 which is said to exceed the number of stars in the universe.21 Scientists have estimated that each of us harbors more than 380 trillion individual viruses living on or inside our bodies,22 most of which do no harm. These difficult-to-comprehend figures are tempered somewhat when one recognizes that a high proportion of viruses in these estimates are copies of the same virus. Nevertheless, estimations of counts of unique viruses are also astounding. For instance, considering just mammals alone, it has been estimated that the earth currently has 5291 unique species of mammals, which in total harbor roughly 40 000 unique species of viruses.23 Approximately 10 000 of these viral species are thought to have potential to cause illness in humans (ie, they are zoonotic).23 Given the large number of diverse viral threats both outside and within a human host, one should marvel that our human immune systems are so effective in preventing viral disease.
Figure 2. The Human Immune System Prevents Most Microbial Assaults From Causing Infection
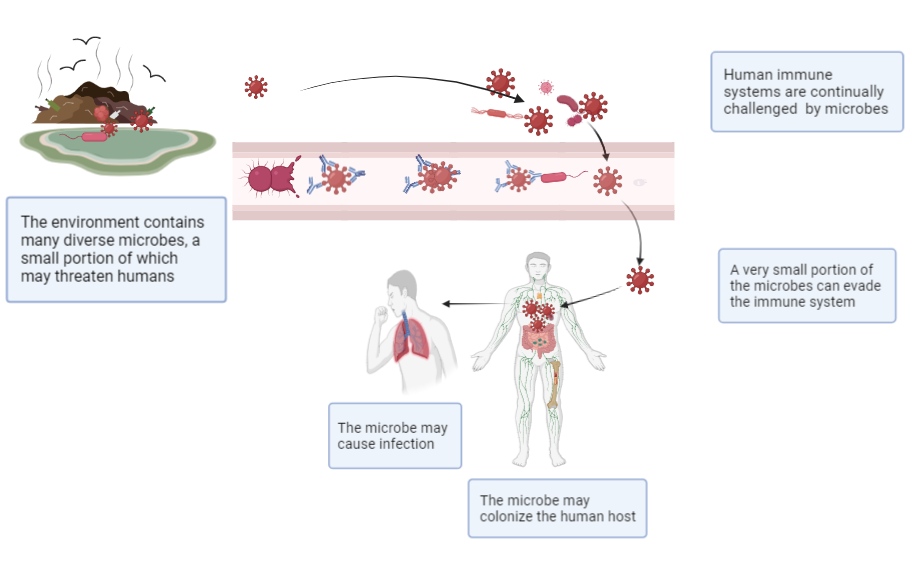
Created with BioRender.com.
Humans live in environments that contain trillions of microbes. Our bodies are extremely effective in preventing infections from the vast majority of these microbes. Nevertheless, a small percentage of these microbes can invade our body and cause disease.
Lesson 2: Extant Medical Diagnostics Can Miss Clinically Important Viral Infections
Modern viral diagnostics are finely tuned to be both sensitive and specific with respect to previously recognized viral pathogens. When previously recognized viruses change or never-before-detected viruses emerge to cause disease, they are often missed by routine clinical diagnostics, as illustrated recently by delays incurred in developing and making widely available viral diagnostics for the A/(H1N1)pdm09 virus24 and some novel emergent coronaviruses. Fortunately, modern laboratory diagnostic science is reducing these delays. For instance, in 2003, it took approximately 5 months for the newly emergent SARS-CoV-1 to be identified and sequenced, such that molecular diagnostics could be developed.25 In contrast, in 2019, SARS-CoV-2 was sequenced approximately 3 weeks after visualization with electron microscopy.25 Nevertheless, in January and February 2020, 6 European laboratories reported delays of up to 2 weeks in implementing molecular diagnostics for SARS-CoV-2 due to contaminated material,26 and by June 2020, these diagnostics were still not widely available in low- and middle-income countries in Africa and South Asia.27 Hence, clinicians are wise to recognize that a negative clinical diagnostic assay—even a multiplex molecular assay—does not necessarily rule out infection from a targeted virus that has changed or a never-before-characterized virus.
Lesson 3: All Viral Spillovers Are Not the Same
In a 2018 report, Carroll et al estimated that there were 1.67 million as-yet-undiscovered viruses in mammal and bird hosts.28 As much of the microbial world is yet to be discovered, we need to develop tools to make those discoveries, especially for microbes that have just spilled over to infect humans but are not yet efficient in causing human-to-human transmission.29 Detecting a pathogen in early spillover and mitigating transmission before it further adapts to humans could prevent human epidemics.29,30 Special surveillance strategies and special broad diagnostic approaches are needed to pick up such viruses before they become problematic.
As there are many possible viral threats, it would be strategic to focus on viruses with the greatest spillover risk to humans. However, rates of virus spillover and spillback are largely unknowable because the vast majority of occurrences are dead-end events.31 Moreover, many zoonotic virus infections might not cause signs or symptoms of disease. Nonetheless, scientific teams have conducted retrospective studies of the small proportion of detected spillover events and have attempted to establish spillover risk scores for specific viruses. One such modeling team has developed a rather compelling ranking system (SpillOver platform) for animal viruses; risk factors were identified on the basis of expert opinion as well as an examination of the limited scientific evidence.32 In ranking the spillover potential of 887 wildlife viruses, Grange et al observed that their model identified 12 known zoonotic wildlife viruses, including SARS-CoV-2, but also ranked a number of other wildlife viruses as having high spillover risk that had not previously been observed to infect humans.32 However, in their critique of this analysis, Wille et al recently argued: “we demonstrate that the virological data on which these analyses are conducted are incomplete, biased, and rapidly changing with ongoing virus discovery.”33 Given these data deficiencies, other groups rely more on expert opinion in evaluating the limited available data and in ranking viruses for risk of either spillover or the generation of human epidemics. For instance, the World Health Organization is ambitiously assembling 20 to 25 groups of experts to review and prioritize viruses in numerous viral families for their epidemic and pandemic risk.34
Such data-centric and expert panel reviews have generated considerable debate on how best to conduct surveillance of spillover- and spillback-prone viruses. Some scientists have embraced a broad viral discovery approach with a central focus on studying the genetic characteristics of wildlife viruses.28 Other scientists argue that surveillance would be better focused on performing One Health-related studies in geographical areas of previous emerging infectious disease risk where humans are in close contact with dense populations of animals.2,29,30,35,36,37 For example, the first author (GCG) and colleagues were fortunate to partner with Chinese scholars in simultaneously studying Chinese pig workers, pigs, and pig farms for evidence of influenza A virus spillover from pigs to humans. We found that immunologically naïve swine workers had markedly increased risk of infection with swine influenza viruses.38
Whatever the logic, many experts agree that surveillance tools should focus on detecting novel viruses from families of viruses at risk of spilling over to humans and causing disease and subsequent epidemics. Some argue, like the first author (GCG) and colleagues’ research team, that respiratory viruses are the highest priority because they spread rapidly and are difficult to control.29,36,39 A number of viral families have a history of causing respiratory disease epidemics: Orthomyxoviridae (contains influenza viruses), Coronaviridae (contains coronaviruses), and Paramyxoviridae (contains parainfluenza viruses).29,36,40 To these we add Adenoviridae (contains adenoviruses), Pneumoviridae (contains respiratory syncytial virus and human metapneumovirus) and Picornaviridae (contains enteroviruses and rhinoviruses), as each of these viral families has been responsible for additional human respiratory virus epidemics.39 Other short lists of important human-disease-causing viral families include Bunyavirales (contains hantavirus), Filoviridae (contains ebolavirus), Flaviviridae (contains dengue and yellow fever viruses), and Togaviridae (contains Sindbid virus).40,41 Recently, Valero-Rello and Sanjuan studied a database of 12 888 virus-host associations (comprising 5149 viruses and 1599 host species) and concluded that enveloped viruses are more likely to spill over to new species.42 No matter the specific viral families included in novel virus surveillance efforts, better viral discovery diagnostics are needed to detect specific viral spillover threats within these viral families such that future human epidemics could be averted.
With awareness that spillover events are not homogeneous, clinicians can help detect spillover events by being vigilant and considering novel zoonotic spillover as a possible cause of unusual signs and symptoms in their patients, especially those with intense occupational exposure to animals. Simply asking the question, “Could my patient be suffering a spillover or spillback novel virus infection?” could be the first step in novel virus discovery.
Lesson 4: Viral Spillback Is a Global and Domestic Public Health Threat
Zoonotic viruses that cause disease in humans can spill over to other animal populations, mutate or recombine, and spill back to humans, causing additional disease outbreaks. While the movement of the A/(H1N1)pdm09 virus from humans to various animal species, especially pigs, has caused considerable alarm,15,43 it is the generation of novel variants of SARS-CoV-2 in these new animal hosts that is of most concern. As of August 23, 2023, human-adapted SARS-CoV-2 had infected at least 34 unique animal species in 39 countries.44,45 Current evidence suggests that SARS-CoV-2 has adapted to new mink and white tail deer hosts and at least some mink-associated novel variants are capable of spilling back to humans.46 Although current evidence suggests that pigs47,48,49,50,51,52,53 and cattle54,55,56 are not routinely susceptible to infection with circulating strains of SAR-CoV-2, were SARS-CoV-2 strains to adapt to infect livestock, other circulating strains of enzootic coronaviruses in livestock might increase the risk of novel virus generation through recombination. When confronted with a patient with unexplained illness and negative diagnostic tests, clinicians are wise to ask questions about a patient’s exposure to animals and to consider animal pathogen involvement in the patient’s signs and symptoms.
Lesson 5: Clinicians Have Obligations to Be Knowledgeable, Vigilant Agents of Detection
We currently only detect a small proportion of spillover events, and often, when we do, it is too late to take public health action. We need to adopt strategies to detect spillover events when and where they arise. Clinicians need to be vigilant and engage public health officials (locally, regionally, and, if necessary, through national or international reporting networks such as PROMED57 and GOARN58) when they have suspicions that the populations they serve might be experiencing spillover or spillback events. While one might argue that the A/(H1N1)pdm09 virus and SARS-CoV-2 spread so rapidly and efficiently that early warning would not have made much of a difference, other spillover events might be more amenable to public health intervention, as the majority of spillover viruses need considerable time to circumvent numerous immunologic barriers36 to fully adapt to their new human host.29 Recently, G.C.G. and colleagues49 and other research teams59,60 detected novel animal coronaviruses among humans with acute respiratory disease in several geographical areas.61 It seems likely that these spillover viruses have not yet sufficiently adapted to their new human hosts to cause efficient human-to-human transmission. Such viral detections afford opportunity for the development of targeted molecular diagnostics to ascertain viral ecology and stop virus transmission. Given appropriate pan-species diagnostics,29,62 a vigilant health care professional might sound an early alarm and engage public health authorities, who might then take measures to mitigate the threat before the virus has fully adapted to humans.
Responding to International and National Vulnerability
Recent epidemics have demonstrated how novel zoonotic respiratory viruses can cause widespread human infections. We have reviewed the important concepts of zoonotic virus spillover and spillback, seeking to inform clinicians of the important roles they may play in detecting novel viruses and helping to reduce such viruses’ spread.
References
-
Lieberman JI, Rodge TJ, Shalala DE, et al. The Athena Agenda: Advancing the Apollo Program for Biodefences. Bipartisan Commission on Biodefense; 2022. Accessed September 15, 2023. https://biodefensecommission.org/wp-content/uploads/2022/04/Athena-Report_v4.pdf
-
Aarestrup FM, Bonten M, Koopmans M. Pandemics—One Health preparedness for the next. Lancet Reg Health Eur. 2021;9:100210.
- O’Leary DR, Marfin AA, Montgomery SP, et al. The epidemic of West Nile virus in the United States, 2002. Vector Borne Zoonotic Dis. 2004;4(1):61-70.
- Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967-1976.
- Towner JS, Khristova ML, Sealy TK, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J Virol. 2006;80(13):6497-6516.
- Dawood FS, Iuliano AD, Reed C, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12(9):687-695.
-
Khan S, El Morabet R, Khan RA, et al. Where we missed? Middle East respiratory syndrome (MERS-CoV) epidemiology in Saudi Arabia; 2012-2019. Sci Total Environ. 2020;747:141369.
- Su S, Gu M, Liu D, et al. Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol. 2017;25(9):713-728.
-
Coltart CEM, Lindsey B, Ghinai I, Johnson AM, Heymann DL. The Ebola outbreak, 2013-2016: old lessons for new epidemics. Philos Trans R Soc Lond B Biol Sci. 2017;372(1721):20160297.
- Musso D, Ko AI, Baud D. Zika virus infection—after the pandemic. N Engl J Med. 2019;381(15):1444-1457.
-
Ortiz-Martínez Y, Patiño-Barbosa AM, Rodriguez-Morales AJ. Yellow fever in the Americas: the growing concern about new epidemics. F1000 Res. 2017;6:398.
- Kafetzopoulou LE, Pullan ST, Lemey P, et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science. 2019;363(6422):74-77.
-
Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. 2020;7(1):11.
-
Porta M, ed; International Epidemiological Association. A Dictionary of Epidemiology. 6th ed. Oxford University Press; 2014.
-
Messenger AM, Barnes AN, Gray GC. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom-documented human biological threats to animals. PLoS One. 2014;9(2):e89055.
-
Regional Office for South-East Asia. A Brief Guide to Emerging Infectious Diseases and Zoonoses. World Health Organization; 2014. Accessed October 26, 2023. https://iris.who.int/bitstream/handle/10665/204722/B5123.pdf?sequence=1&isAllowed=y
-
Shapiro JT, Víquez RL, Leopardi S, et al. Setting the terms for zoonotic diseases: effective communication for research, conservation, and public policy. Viruses. 2021;13(7):1356.
-
One Health Initiative Task Force. One Health: A New Professional Imperative. American Veterinary Medical Association; 2008. Accessed October 27, 2023. https://www.avma.org/sites/default/files/resources/onehealth_final.pdf
- Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96(5):2192-2197.
-
Mushegian AR. Are there 1031 virus particles on earth, or more, or fewer? J Bacteriol. 2020;202(9):e00052-20.
-
Microbiology by numbers. Nat Rev Microbiol. 2011;9(9):628.
-
Ma ZS. Spatial heterogeneity analysis of the human virome with Taylor’s power law. Comput Struct Biotechnol J. 2021;19:2921-2927.
- Carlson CJ, Zipfel CM, Garnier R, Bansal S. Global estimates of mammalian viral diversity accounting for host sharing. Nat Ecol Evol. 2019;3(7):1070-1075.
-
Baldo V, Bertoncello C, Cocchio S, et al. The new pandemic influenza A/(H1N1)pdm09 virus: is it really “new”? J Prev Med Hyg. 2016;57(1):E19-E22.
-
Mardian Y, Kosasih H, Karyana M, Neal A, Lau CY. Review of current COVID-19 diagnostics and opportunities for further development. Front Med (Lausanne). 2021;8:615099.
- Mögling R, Meijer A, Berginc N, et al. Delayed laboratory response to COVID-19 caused by molecular diagnostic contamination. Emerg Infect Dis. 2020;26(8):1944-1946.
-
Peplow M. Developing countries face diagnostic challenges as the COVID-19 pandemic surges. Chemical & Engineering News. June 26, 2020. Accessed January 20, 2023. https://cen.acs.org/analytical-chemistry/diagnostics/Developing-countries-face-diagnostic-challenges/98/i27
- Carroll D, Daszak P, Wolfe ND, et al. The Global Virome Project. Science. 2018;359(6378):872-874.
-
Gray GC, Robie ER, Studstill CJ, Nunn CL. Mitigating future respiratory virus pandemics: new threats and approaches to consider. Viruses. 2021;13(4):637.
-
Gray GC, Abdelgadir A. While we endure this pandemic, what new respiratory virus threats are we missing? Open Forum Infect Dis. 2021;8(3):ofab078.
-
Ellwanger JH, Chies JAB. Zoonotic spillover: understanding basic aspects for better prevention. Genet Mol Biol. 2021;44(1)(suppl 1):e20200355.
-
Grange ZL, Goldstein T, Johnson CK, et al; Expert Panel; PREDICT Consortium; University of Edinburgh Epigroup members those who wish to remain anonymous. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc Natl Acad Sci U S A. 2021;118(15):e2002324118.
-
Wille M, Geoghegan JL, Holmes EC. How accurately can we assess zoonotic risk? PLoS Biol. 2021;19(4):e3001135.
-
WHO to identify pathogens that could cause future outbreaks and pandemics. News release. World Health Organization; November 21, 2022. Accessed September 15, 2023. https://www.who.int/news/item/21-11-2022-who-to-identify-pathogens-that-could-cause-future-outbreaks-and-pandemics
- Vora NM, Hannah L, Lieberman S, Vale MM, Plowright RK, Bernstein AS. Want to prevent pandemics? Stop spillovers. Nature. 2022;605(7910):419-422.
- Holmes EC. The ecology of viral emergence. Annu Rev Virol. 2022;9(1):173-192.
- Holmes EC, Rambaut A, Andersen KG. Pandemics: spend on surveillance, not prediction. Nature. 2018;558(7709):180-182.
- Borkenhagen LK, Wang GL, Simmons RA, et al. High risk of influenza virus infection among swine workers: examining a dynamic cohort in China. Clin Infect Dis. 2020;71(3):622-629.
-
Bailey ES, Fieldhouse JK, Choi JY, Gray GC. A mini review of the zoonotic threat potential of influenza viruses, coronaviruses, adenoviruses, and enteroviruses. Front Public Health. 2018;6:104.
-
Moisi A, Thomas L, Williams G; CSIRO Futures. Strengthening Australia’s Pandemic Preparedness: Science and Technology-Enabled Solutions. CSIRO; 2022. Accessed October 27, 2023. https://www.csiro.au/-/media/Services/Futures/Strengthening-Australias-pandemic-preparedness/22-00161_SER-FUT_REPORT_StrengtheningPandemicPreparedness_WEB_220527.pdf
-
National Institute of Allergy and Infectious Diseases. NIAID pandemic preparedness plan. National Institute of Allergy and Infectious Diseases; 2021. Accessed October 26, 2023. https://www.niaid.nih.gov/sites/default/files/pandemic-preparedness-plan.pdf
-
Valero-Rello A, Sanjuán R. Enveloped viruses show increased propensity to cross-species transmission and zoonosis. Proc Natl Acad Sci U S A. 2022;119(50):e2215600119.
- Nelson MI, Vincent AL. Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface. Trends Microbiol. 2015;23(3):142-153.
-
SARS-ANI VIS: a global open access dataset of reported SARS-CoV-2 events in animals. Complexity Science Hub Vienna. Updated August 23, 2023. Accessed July 23, 2023. https://vis.csh.ac.at/sars-ani/#infections
-
Nerpel A, Yang L, Sorger J, Käsbohrer A, Walzer C, Desvars-Larrive A. SARS-ANI: a global open access dataset of reported SARS-CoV-2 events in animals. Sci Data. 2022;9(1):438.
-
Pappas G, Vokou D, Sainis I, Halley JM. SARS-CoV-2 as a Zooanthroponotic infection: spillbacks, secondary spillovers, and their importance. Microorganisms. 2022;10(11):2166.
- Meekins DA, Morozov I, Trujillo JD, et al. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):2278-2288.
- Sikkema RS, Tobias T, Oreshkova N, et al. Experimental and field investigations of exposure, replication and transmission of SARS-CoV-2 in pigs in the Netherlands. Emerg Microbes Infect. 2022;11(1):91-94.
- Vergara-Alert J, Rodon J, Carrillo J, et al. Pigs are not susceptible to SARS-CoV-2 infection but are a model for viral immunogenicity studies. Transbound Emerg Dis. 2021;68(4):1721-1725.
- Schlottau K, Rissmann M, Graaf A, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020;1(5):e218-e225.
-
Haddock E, Callison J, Seifert SN, et al. Three-week old pigs are not susceptible to productive infection with SARS-COV-2. Microorganisms. 2022;10(2):407.
-
Happi AN, Ayinla AO, Ogunsanya OA, et al. Detection of SARS-CoV-2 in terrestrial animals in Southern Nigeria: potential cases of reverse zoonosis. Viruses. 2023;15(5):1187.
-
Opriessnig T, Huang YW. SARS-CoV-2 does not infect pigs, but this has to be verified regularly. Xenotransplantation. 2022;29(5):e12772.
-
Fiorito F, Iovane V, Pagnini U, et al. First description of serological evidence for SARS-CoV-2 in lactating cows. Animals (Basel). 2022;12(11):1459.
- Wernike K, Bottcher J, Amelung S, et al. Antibodies against SARS-CoV-2 suggestive of single events of spillover to cattle, Germany. Emerg Infect Dis. 2022;28(9):1916-1918.
- Ulrich L, Wernike K, Hoffmann D, Mettenleiter TC, Beer M. Experimental infection of cattle with SARS-CoV-2. Emerg Infect Dis. 2020;26(12):2979-2981.
-
Dyer O. Global disease reporting service ProMED faces uncertain future as funding runs out. BMJ. 2023;382:p1872.
-
Christensen R, Fisher D, Salmon S, Drury P, Effler P. Training for outbreak response through the Global Outbreak Alert and Response Network. BMC Med. 2021;19(1):123.
- Vlasova AN, Diaz A, Damtie D, et al. Novel canine coronavirus isolated from a hospitalized pneumonia patient, East Malaysia. Clin Infect Dis. 2021;74(3):446-454.
- Vlasova AN, Toh TH, Lee JS, et al. Animal alphacoronaviruses found in human patients with acute respiratory illness in different countries. Emerg Microbes Infect. 2022;11(1):699-702.
-
Lednicky JA, Tagliamonte MS, White SK, et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. MedRxiv. 2021;2021.03.19.21253391.
-
Xiu L, Binder RA, Alarja NA, et al. A RT-PCR assay for the detection of coronaviruses from four genera. J Clin Virol. 2020;128:104391.